Sequencing DNA from Individual Cells Yields Dramatic New Information
November 1, 2012
Image: Visuals Unlimited/Corbis
Nicolas Navin wanted to work out the sequence from individual cancer cells to see how they had mutated and diverged as the cancer grew. Back in 2010, he was a postdoctoral fellow at Cold Spring Harbor Laboratory in New York and exploring the genetic changes that occur during breast cancer.
Cells like to stick together and no matter how hard he tried, he could never ensure that he had a single cell to work with. Eventually, he started using chemicals to dissolve the outer layers of cells and release the nuclei. Then, the nuclei were separated using an automated cell sorter and their DNA was extracted. This process was repeated for 100 cells and the obtained sequences revealed how the tumor had evolved from a few rogue cells into a complex mixture of genetically distinct ones.
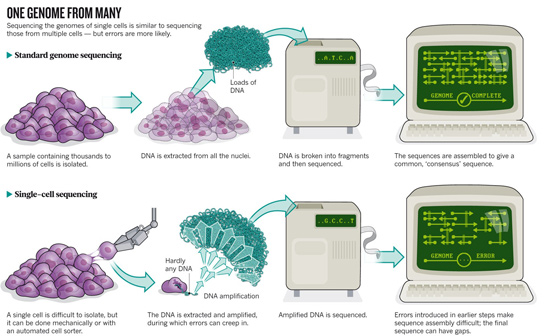
This process would have been unthinkable a decade ago, and it’s still a remarkable feat. Technology has dramatically reduced costs and made genome sequencing routine. However, most human genomes are sequenced from DNA extracted from multiple cells, which misses the differences between cells that could be crucial to controlling gene expression, cell behavior and drug response.
Infographic from Nature
Amplifying the amount of DNA in a single cell until there’s enough to sequence without introducing too many errors is still difficult. Bioinformatics are required to stitch together the data and deal with artifacts. For this reason, research groups have started with cells that are easily separable, like sperm, or those that are likely to have dramatic genomic differences, like tumor cells.
One of the most interesting facets of this research is how tissues and organs manage to work coherently despite the differences between cells. In his first effort, Navin was able to sequence only about 10% of the DNA, which wasn’t enough to see individual mutations, but good enough to study larger segments that are commonly duplicated or deleted.
Tumors are made up of three major populations of cells, which emerge from the root tumor cell population at different times. Since Navin moved to Texas, he’s started a research group focused entirely on single-cell genomics. He can now piece together 90% of a cell’s genome, which allow him to study the mutations in individual cancer cells in much more detail.
Sequencing a tumor as a whole yielded six mutations in the cancer-associated genes. When the scientists sequenced four individual cells, they identified hundreds of additional mutations, many of which were unique or private to that individual cell. The team found that about 1% of mutations, involving 12,000 to 20,000 base pairs, differed from cell to cell and that these variations could not be detected when the cells were sequenced together.
The cost of sequencing individual cells was about $1,000 per cell. Timour Baslan, Navin’s former colleague, is working on ways to bring down this cost. They add genetic barcodes to a cell’s DNA, allowing them to sequence cell genomes en masse then identify the sequences from individual cells. Using these barcodes, with improved bioinformatic approaches, brings the cost of sequencing a cell’s genome down to about $60.
Baslan and his colleagues are using the techniques to study which tumor cells are left after chemotherapy and why these cells are resistant. This could guide future treatments. By sequencing a few cells, scientists might be able to get an idea of the heterogeneity in a tumor before chemotherapy, and this could affect the choice of the agent to use in the treatment.
Adam Auton, statistical geneticist at Albert Einstein College of Medicine, in New York, is using sperm to study recombination. Recombination is one of the fundamental forces that shapes genetic diversity, states Auton.
Working with a team from the Chinese sequencing company BGI, Auton sequenced nearly 200 sperm cells and was able to estimate the recombination rate for the man who had donated them. There were an average of 24.5 recombination events per sperm cell, which is in line with estimates from indirect experiments. The location of these recombination hotspots could help population biologists map the position of the genetic variants associated with disease.
No comments:
Post a Comment