Genes Related to Host-Microbe Interactions Linked to IBD
November 5, 2012
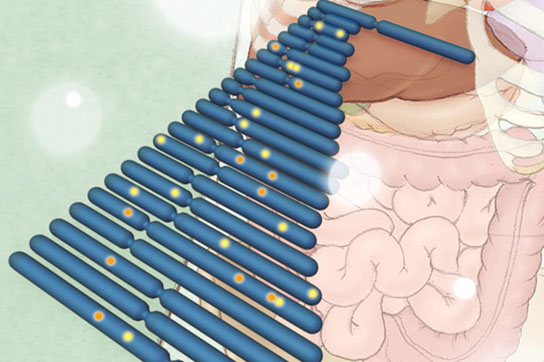
Researchers have identified more than 70 new genetic risk factors (shown as bright spots on chromosomes, shaded in blue) tied to inflammatory bowel disease (IBD). Image courtesy of Janet Iwasa and Bang Wong
A newly published study identified 71 additional genetic associations for IBD and suggests a strong overlap between IBD susceptibility genes and genes tied to the immune system’s response to mycobacterial infections, including tuberculosis and leprosy.
Crohn’s disease (CD) and ulcerative colitis (UC) — inflammatory diseases of the gastrointestinal tract — have puzzled the scientific community for decades. Ten years ago, researchers recognized that both genes and environment contributed to these diseases but knew little about precisely how and why illness occurred.
Today, researchers from across the CD and UC communities have come together to share raw data as well as newly collected genetic information to dissect the biology of a group of conditions that affect millions of people worldwide. Their research centers on the two diseases, which are collectively known as inflammatory bowel disease (IBD), and suggests a fundamental connection between risk of IBD and genes involved in other immune-related diseases and the immune system’s response to pathogens. The work by researchers from the Broad Institute of Harvard and MIT, Harvard-affiliated Massachusetts General Hospital (MGH), Yale School of Medicine, Cedars-Sinai Medical Center, and dozens of other organizations appears in a Nature paper this week.
“This study marks the first time we’ve acquired and combined the raw data from so many research studies around the world and also the first time we’ve jointly analyzed Crohn’s with ulcerative colitis,” said Mark Daly, one of the senior authors of the work and senior associate member of the Broad Institute and co-director of its Program in Medical and Population Genetics. Daly is also chief of the Analytic and Translational Genetics Unit at MGH and an associate professor at Harvard Medical School (HMS). “We’ve been able, with this study, to evaluate the evidence for both diseases simultaneously, and discovered that the majority of genetic risk factors are associated with both diseases.”
“There’s been a paradigm shift in our understanding of IBD. This gene discovery process offers an opportunity to begin identifying new targets for treatment, better diagnostic tools, and, in the long term, personalized care for patients,” said co-author HMS Associate Professor of Medicine Ramnik Xavier, a senior associate member of the Broad Institute, and chief of gastroenterology and director of the Center for the Study of Inflammatory Bowel Disease at MGH. “We now have the necessary starting material to understand the pathways that contribute to Crohn’s disease and ulcerative colitis, and we also have a framework to better appreciate that they may not be two distinct diseases, but rather collections of many different diseases.”
Crohn’s disease and ulcerative colitis have much in common: Both cause many of the same gastrointestinal symptoms and both are marked by an improper response by the body’s immune system to harmless cells or bacteria. Over the past 10 years, researchers have performed genomewide association studies, looking across the genomes of thousands of patients with either CD or UC and comparing them with genomes from people who do not have these diseases to find significant genetic differences. The new study not only brings together the original data from those previous analyses, but also adds genetic information from another 40,000 people either with or without a form of IBD.
“If we want to get more hits but also dissect the differences between Crohn’s disease and ulcerative colitis or understand the commonalities, we really need to share all of our genetic data,” said co-first author Stephan Ripke, a researcher at the Broad Institute and MGH. Ripke worked closely with co-first author Luke Jostins of the Sanger Institute to combine and then analyze genetic information collected by researchers from many different institutions.
The new study identified 71 additional genetic associations for IBD, many of which have been previously implicated in other immune-related disorders, including ankylosing spondylitis and psoriasis. The new research also suggests a strong overlap between IBD susceptibility genes and genes tied to the immune system’s response to mycobacterial infections, including tuberculosis and leprosy. Researchers have observed similarities between the immune response in CD and that seen in tuberculosis, and they hypothesize that CD could be an aberrant response to certain harmless organisms present in the gut that trigger a similar reaction.
In addition to drawing upon original data from previous studies, the work utilizes a relatively new tool known as the immunochip, which samples 200,000 sites in the genome previously tied to autoimmune and inflammatory diseases.
“As a research community, we designed this array to target genes involved in immune-mediated diseases of all kinds,” said Daly. “This is one of our first studies using the immunochip, and there are many more results that will emerge from these studies — within IBD and across immune diseases.”
At the Broad Institute, researchers are already pursuing some of the pathways identified in previous studies of UC and CD, including autophagy, a process in which infected cells eat themselves to combat microbes.
“This work gives us additional leads to pursue,” said Xavier. “It gives us an opportunity for high-quality, translational research, allowing us to identify core pathways involved in IBD and better understand how genes interact with each other and with the environment, illuminating new pathways that contribute to disease.”
Source: Haley Bridger, Broad Institute Communications; Harvard Gazette
Image: Janet Iwasa and Bang Wong
No comments:
Post a Comment